Last summer, the CASP (Critical Assessment of Structure Prediction) and CAPRI (Critical Assessment of PRedicted Interactions) communities teamed up for a joint round of predictions.
25 targets were made available to both communities in order to predict the 3D structure of those complexes.
These were mostly homodimers with a few tetramers and some heteromers. This meant effectively modelling the structures of the monomers and their assemblies from sequence information only.
The HADDOCK team participated to the server and scoring competitions, using mainly symmetry restraints with the ab-initio docking mode of HADDOCK (based on center-of-mass restraints). We chose not to use any bioinformatics prediction and participate only as server for the prediction since the biological relevance of those complexes was uncertain. In a number of cases, when a template of the dimer was available, we directly modelled the complex by homology modelling and refined it throught the refinement interface of the HADDOCK web server.
The results of this joint CASP/CAPRI prediction round were presented by Shoshana Wodak at the CASP meeting in Cancun, Mexico last December.
The results show that HADDOCK is maintaining its leading position in this field, with the server ranking at the top of all servers (and above pretty much all CASP predictors!).
In the scoring experiment, the HADDOCK score (based on the OPLS force field of Jorgensen) performed extremely well, ranking at the top among all participating CASP and CAPRI groups. This is summarized in the following slide taken from Shoshana Wodak’s presentation:
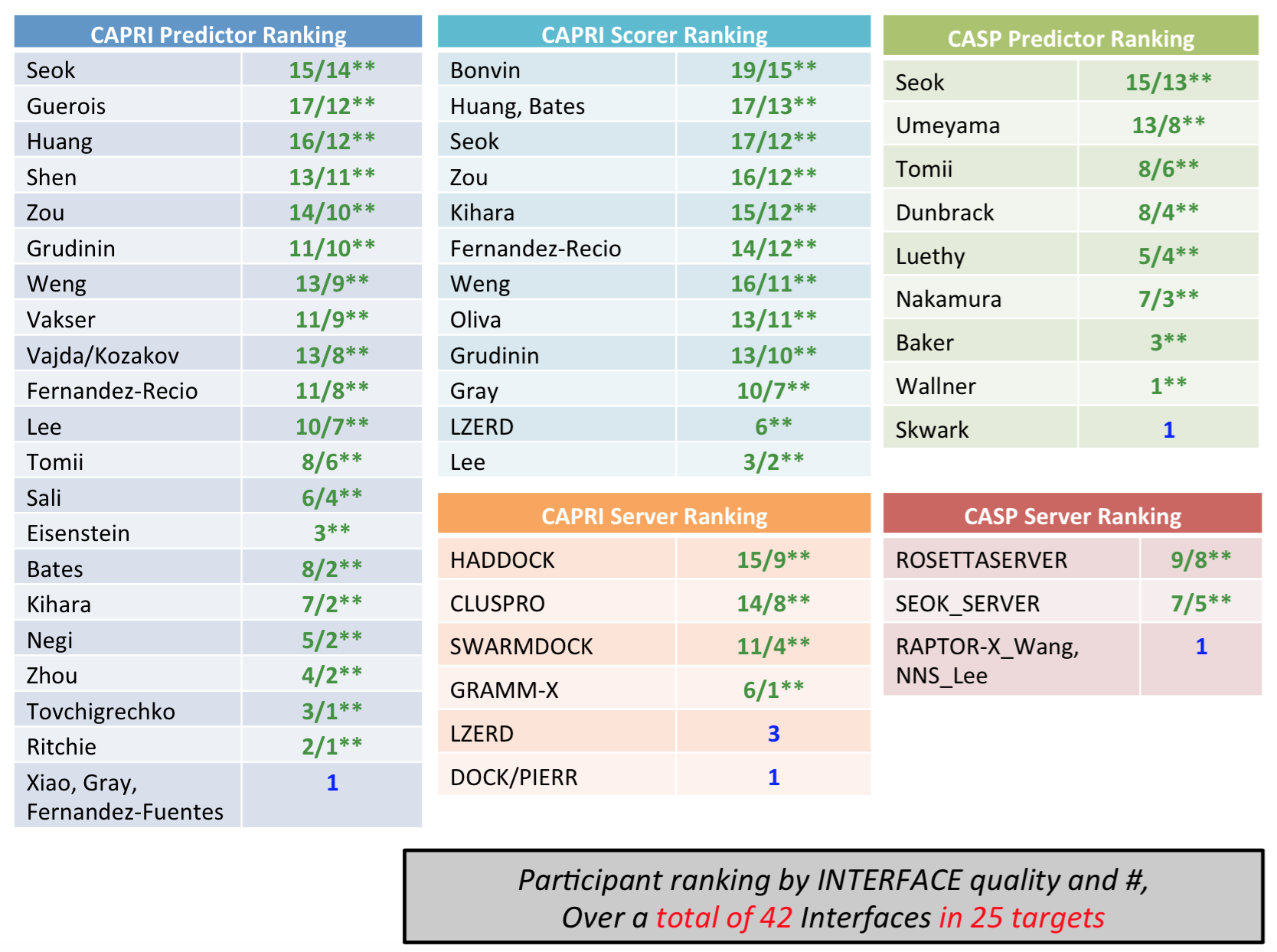
The slides of the full presentation given at the Cancun meeting are available here.
